When I saw the press release about this research (here it is) I wasn’t sure that I could make a column out of it, so I seized on the “big molecule” angle. Now that it’s in print, I’m still not sure I can make a column out of it. But it’s Tuesday – I had to file something!
This is an embarrassing admission for a science fan to make, but it never really occurred to me until recently that molecules can be different sizes.
I know different molecules can contain very different numbers of atoms, but somehow that never affected my impression of how physically large they are. I guess I envisioned all the atoms being smooshed together into one teeny-tiny point that didn’t actually have any size.
That image, which may help explain my chemistry grades, was gently but firmly put aside recently by HarishVashisth, assistant professor of chemical engineering at UNH, when I talked to him about recent research targeting the agricultural pest known as roundworms or nematodes.
Vashisth cares a lot about molecular size because his lab specializes in studying how small-molecule drugs can be used.
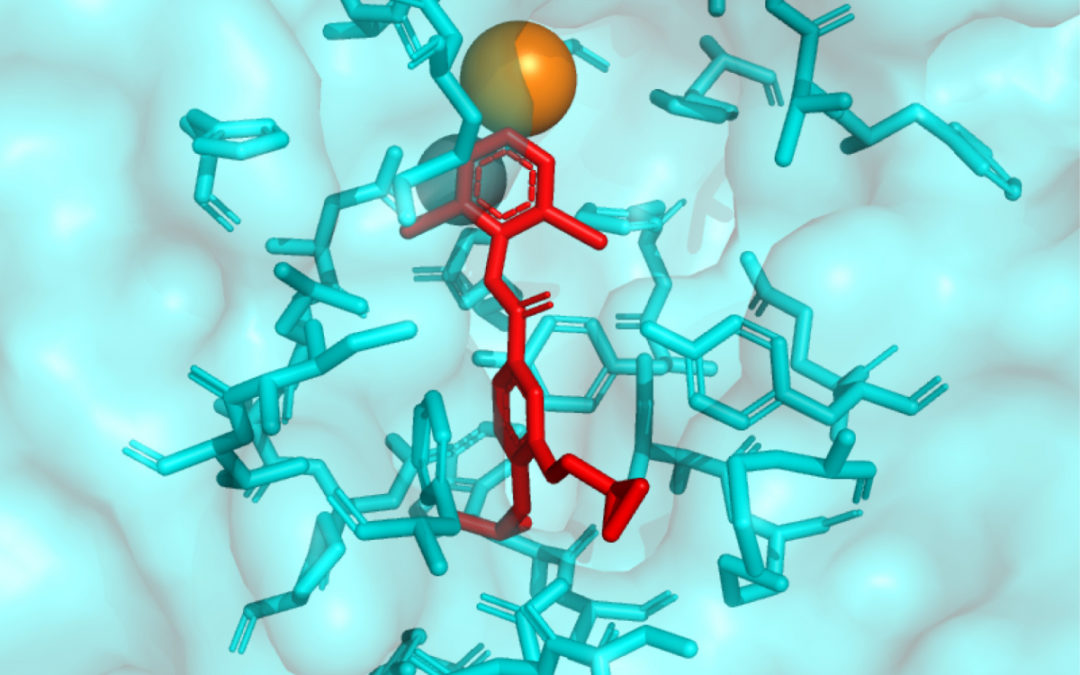
“A very large molecule cannot go across the membrane and attack something inside the cell. A small molecule can sneak through the spaces in the wall and go inside the cell and find small space on the target,” Vashisth said. “It’s very strategic and extremely important to be able to design smaller molecules to attack bigger molecules and get into the cell.”
“Large molecules can be like spheres,” said Vashisth. “There are hills and valleys on the surface – some places that are deeper, other places that are not. … Smaller molecules, they can go and fit into smaller spaces that a large molecule can’t.”
The goal isn’t to blow up the big molecule, only to change what it does.
The big molecules in question are frequently proteins and enzymes which, of course, make much of life work. If you can find little molecules that will alter the actions of proteins and enzymes, then you can do all sorts of things. You might create specialized medicine for us humans or, as in the case we’re talking about here, you might create more environmentally friendly pesticides.
Pesticides are needed because nemadotes are a big problem in agriculture. They’re a class of tiny worms that can infest the roots of many crops and caused literally billions of dollars worth of damage.
UNH has just announced a breakthrough is understanding of phosphodiesterase enzymes or PDEs in nematodes, to better understand how to create PDE inhibitors – small molecules that could interfere with those big enzyme molecules and do such things as make it impossible for nematodes to find food.
Trying to determine how all the PDEswork and then decide which would be a good target for disruption is a huge task that might take decades, said Vashisth. So UNH pursued a shortcut, leading to a recent publication in the online PLOS ONE journal and a patent-pending discovery (available for licensing through UNHInnovation, the university notes).
“Many of these large molecules, like enzymes, are found across species – you find them in mice, find them in humans” and in nematodes, he said. “Since we know how the human enzyme works, we use it as a template.”
They superimposed the genetic specifics of nematodes on the template “got a 3-dimensional model as a first guess.” These models are mostly in computers, but they also 3D print some to assist in analysis.
“There are drugs on the market that attack and block human enzymes. … We took human drugs to see if they will fit in the same place, we physically took the model and tested it,” he said.
Eventually, he said, they found a static model that worked, leapfrogging years of effort. But there’s still a lot to do.
For one thing, molecules move. A static model doesn’t tell you everything you know – that’s where computer modeling, using various UNH supercomputers to perform simulations, is key.
“That captured the physics of how the smaller molecule and bigger molecule talk to each other. We let the model evolve and watched what happens. … This gives us insight in coming years as to how to use this information to design new drugs that are more effective,” he said.
Designing this new drug to battle root-seeking nematodes quires searching through the vast library of available drugs to see which fit into the static model. This is where the modeling really comes into play.
“Maybe out of let’s say 100,000 we might find, say, 50 that bind to our protein. Then we can build dynamic models of for each of them and rank them as to which is more effective than others. Once you have a good ranking you might go with your top 5 and try to do an experiment – inject into nematode and see if it kills the bug in some way,” he said.
In other words, even with the shortcut years of work are ahead. But at least progress is feasible, whereas setting up experiments to test hundreds or thousands of potential drugs wouldn’t be.
Other researchers involved in this work include Prof. Rick Cote, a researcher with the New Hampshire Agricultural Experiment Station; Kevin Schuster, doctoral student in biochemistry; MohammadjavadMohammadi, doctoral student in chemical engineering; Karyn Cahill and Suzanne Matte, former research staff; and Alexis Maillet, undergraduate student in biomedical sciences.

 Return to the Concord Monitor
Return to the Concord Monitor